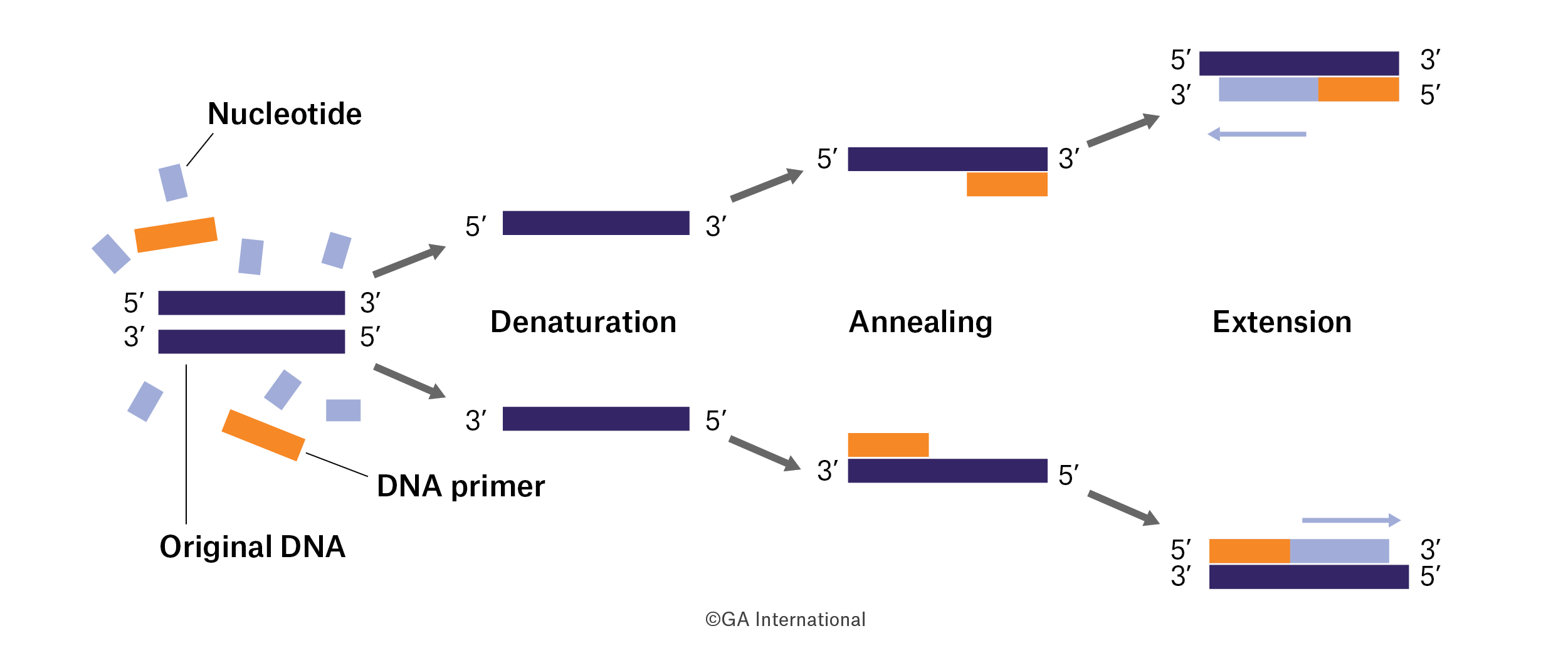
The initial discovery of PCR
The structure of DNA was originally elucidated in the 1950s by Watson and Crick, but it wasn’t until the 1980s that scientists were able to amplify specific DNA sequences.1 The idea for PCR was first devised by Kary Mullis in his now famous drive across Highway 128 in his Honda Civic, in which he had to pull over onto the shoulder of the road once the notion of PCR finally came to him.2 While working for the Cetus Corporation as a chemist in the 1980s, he wanted to design a way to synthetize oligonucleotides as fast or faster than the automated synthesizers that had come out around that time.3 His concept was based off previously existing technologies, including the synthesis of DNA using polymerases. Mullis’ innovation was to use two primers on opposite DNA strands to reproduce the sequence between them, and not just once, but many times over. The first iteration used a DNA polymerase from the bacteria Escherichia coli; unfortunately, this meant that heat would denature both the DNA strands, which was useful, as well as the enzyme, which was not. Using polymerase from E. coli meant that you had to re-add enzyme after each cycle. After some experimentation, Mullis began using the polymerase from Thermophilus aquaticus, a bacterium that can resist extremely high temperatures. Its polymerase was ultimately purified and added to Mullis’ PCR reaction, removing the need to add enzyme after each cycle and making it possible to use even higher temperatures, which made primer binding more stringent and eliminated nonspecific PCR products.2
Ultimately, Kary Mullis would win the Nobel Prize in 1993 for his invention. Roche, which bought out the Cetus Corporation, would also patent it, but not without some controversy. H.G. Khorana challenged the patent rights of PCR based on his work in the 1960s and 1970s, where he developed a “repair replication” method that consisted of steps similar to PCR, including primer annealing, template extension, DNA denaturation, and primer re-annealing.2 Rather than patent the work, Dr. Khorana wanted the public to have free access to his discovery. Very few knew about his method though, as the technologies to create primers and recombinant enzymes in the 1970s were inefficient and expensive, making his technique impractical at the time. Others have also challenged the basis of the patent. Some claim that the patent infringes on the institutions that wish to use PCR for the advancement of knowledge and not for financial gain, while companies like Promega have rendered many aspects of the patent unenforceable in the United States, Australia, and Europe.4
The advent of RT-PCR
PCR wasn’t originally tailored for precise DNA quantification. As the reaction plateaus, exponential amplification of the target sequence stops, and very little product is generated. So, two reactions may start with different copy numbers of DNA but will end up with around the same amount of product. It wasn’t until 1993 that Russ Higuchi, working out of the Cetus Corporation, decided to use ethidium bromide to produce a fluorescent signal that could be monitored in real time as the DNA was being amplified. Others, such a Bob Watson, Carl Witwer, and Kirk Ririe, fine-tuned the technique by creating thermal cyclers specifically designed to monitor PCR-generated fluorescence and to make RT-PCR reactions more efficient.5 Two of the most popular versions of RT-PCR today are the TaqMan assay (named after Pac-Man) and SYBR Green fluorescence.5 TaqMan takes advantage of the DNA polymerase as it nicks the end of a fluorescent probe bound to the target site, releasing it so it can fluoresce, whereas SYBR Green simply binds to double-stranded DNA, amplifying its fluorescent signal as more and more DNA is synthesized.
Digital PCR: a new way of quantifying DNA
The first study that used digital PCR was published in 1988. It wasn’t called digital PCR though, as Saiki et al had not formally named their discovery. In their study, they diluted DNA samples from genomes containing the β-globin gene into a series of samples where β-globin genes had been removed and showed that PCR could be performed with only 1 molecule of DNA. Single molecule PCR was eventually termed “limiting dilution PCR” in the 1990s, with the first use of the technique to quantify the expression of a target gene occurring in 1990. Here, Simmonds et al were able to assess the number of HIV provirus molecules in patient samples using multiple replicates with only a single molecule of DNA in each well and Poisson distribution analysis (which models the number of times an event is likely to occur). However, by the 2000s, fewer and fewer groups were using the technique. In 2007, though, there was a large uptick in the number of publications, mostly in engineering and microfluidics journals, using the technique under the name digital PCR.6
PCR is now one of the most widely used biochemical techniques. It’s employed by clinicians to screen for viral and bacterial infections, to detect cancer, and to diagnose genetic diseases. Biomedical research also depends on PCR to accurately quantify both RNA and DNA in samples, to design expression vectors and clone genes, and to genotype mice and other animals. The technique has come a long way since the 1980s (or the 1970s), with new enzymes and reagents available that make it more efficient and precise, becoming a technological staple in hospitals, research institutes, schools (no high school biology class is complete without learning about PCR), and forensic labs.
LabTAG by GA International is a leading manufacturer of high-performance specialty labels and a supplier of identification solutions used in research and medical labs as well as healthcare institutions.
References:
- Kaunitz JD. The Discovery of PCR: ProCuRement of Divine Power. Dig Dis Sci. 2015;60(8):2230-2231.
- Bartlett J, Stirling D. A Short History of the Polymerase Chain Reaction. In: Humana Press Inc., ed. PCR Protocols. Totowa, NJ; 2003:3-6.
- Perkel J. PCR: Past, Present, & Future. Sci. 2013:1-6.
- Carroll, P; Casimir D. PCR Patent Issues. In: Inc. HP, ed. PCR Protocols. Totowa, NJ; 2003:7-14.
- Williams PM. The beginnings of real-time PCR. Clin Chem. 2009;55(4):833-834.
- Morley AA. Digital PCR: A brief history. Biomol Detect Quantif. 2014. doi:10.1016/j.bdq.2014.06.001